-
30
Jan
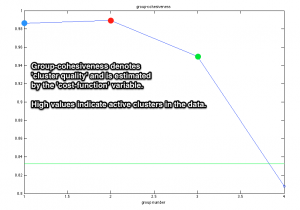
The decision about the actual number of active neurons is an open issue in spike sorting, with sparsely firing neurons and background activity the most influencing factors. Dominant-sets clustering algorithm is a graph-theoretical algorithmic procedure that successfully addresses this issue. The quality of grouping in the data is evaluated with the estimation of ‘cohesiveness’, i.e. a cluster-quality measure, for each group.
Remarks:
- In this example, clustering will be applied on the coordinates of the data (spikes) in ISOMAP space.
- Any other multidimensional coordinates/features (even raw waveforms) may be used.
- Results will not be identical each time, as the algorithm is randomly initialized when approaching the adjacency matrix of the graph.
To reproduce this tutorial in MATLAB you will need :
1. Memo script for MATLAB and sample data to reproduce the results shown below.
In this tutorial we will use simulated spikes from 3 neurons, one being a sparsely-firing one.
Select the data (Feature extraction has been initially applied using ISOMAP algorithm)
For details, see here: Using ISOMAP algorithm for feature extraction in spike sorting
[rx,ry]=min(R);
REDUCED_DIMENSIONALITY=ry;
X_data=Y.coords{REDUCED_DIMENSIONALITY}’;
Build the similarity weighted adjacency matrix first
Estimate Euclidean inter-point distances
d=(dmatrix(X_data));
Transform distances to similarity weights.
factor=5;
sigma=factor*mean(mean(d));
‘A’ is the similarity (weighted adjacency) matrix
clear A; A=exp(-d/sigma); A=A-diag(diag(A));
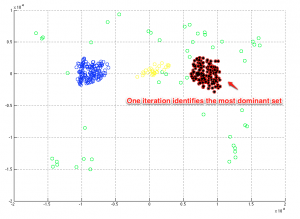
Let us apply one iteration of the dominant-sets algorithm (and look for the most dominant set)
[sel_list,rest_list,ordered_list,memberships,cost_function] = dominant_set_extraction(A);
The most dominant set is marked in black color. Let us proceed to the complete solution now.
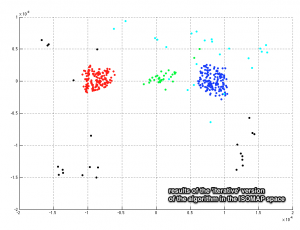
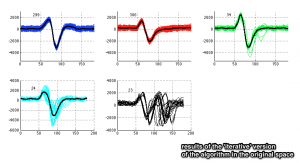
Apply the dominant-sets algorithm iteratively.
[groups,no_groups,cost_function,f_ini] = iterative_dominant_set_extraction(A);
Notes
- False positives in the green cluster have lowered its cohesiveness value, in comparison to the other clusters.
- ‘sigma’ (σ) is a real positive number reflecting the ‘radius of influence’ algorithm which it controls the clustering resolution. A high value -> under-clustering, while a low value -> over-clustering. For details see [1].
- Optimum values of ‘sigma’ may be investigated using a grid optimization process, or empirically based on experimental data (as in [1]). Here, for the sake of proximateness, we use a simpler approach based on ‘d’ matrix. In this case, altering ‘factor’ values affects the output of the algorithm. (Typical values in [2,6] seem to work well.)
If you find this tutorial useful, please cite:
[1] Adamos DA, Laskaris NA, Kosmidis EK, Theophilidis G, “In quest of the missing neuron: Spike sorting based on dominant-sets clustering“. Computer Methods and Programs in Biomedicine 2012, vol.107 (1), pp.28-35. | http://dx.doi.org/10.1016/j.cmpb.2011.10.015
For more information see: http://neurobot.bio.auth.gr/spike-sorting/
(C) D.A. Adamos, 2012.
- Published by Dimitrios A. Adamos in: Software Tutorials
- RSS feed subscription!

 Neurobot via RSS
Neurobot via RSS
